ウェブサイト:
Nanodigmbio
ウェブサイト:
Nanodigmbio
カタログの抜粋

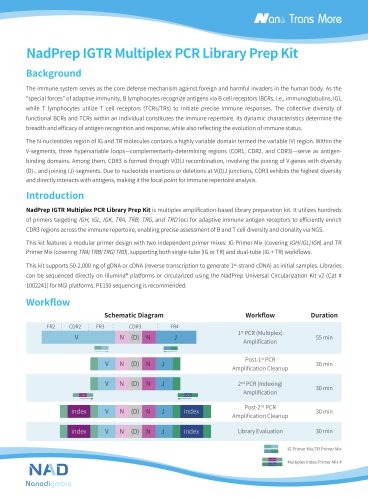
OncoFu Elite (for RNA) Panel Background Fusion gene plays an important role as an oncogenic driver in many cancer types. The most common fusion gene targets in solid tumors are tyrosine kinases, such as ALK, ROS1, and RET, which may cause abnormal sequences, functional protein production, or dysregulation of gene expression, thereby causing or promoting tumorigenesis. A number of targeted drugs targeting the corresponding fusion genes such as ALK, RET, ROS1, FGFRs, and NTRKs have been approved for clinical use. Targeted inhibitors of multiple fusion gene targets have significant therapeutic effects in the clinical treatment of tumor and are the main force in the precision medicine . Therefore, the detection of fusion genes is of great significance for clinical diagnosis, treatment, and prognosis. At present, the conventional methods of fusion gene diagnosis mainly include fluorescence in situ hybridization (FISH), quantitative real-time PCR (RT-PCR), and IHC. Generally, these methods have low resolution and throughput, can only detect a single fusion gene, and cannot identify new fusion gene partners or analyze complex structural reorganizations. Besides, these methods greatly rely on the judgment experience of the technicians and have limited expansion capacity of application. In recent years, DNA-based targeted capture sequencing technology (DNACap) has been used more and more in clinical testing laboratories because of its various advantages such as independence on prior knowledge of fusion, the ability to discover new fusion subtypes, and high sensitivity. However, DNACap also has certain limitations in the detection of fusion genes, and it is not technically feasible when there are repetitive sequences in the fusion-related region or the intron span is too large. In comparison, RNACap has the following advantages: The intron sequence in mRNA has been cleaved, and the reduction of the target regions improves the breadth and sensi- tivity of fusion analysis; Targeted enrichment technology focuses on pivotal fusion-related genes, and RNA library preparation does not require the procedure of ribosomal RNA removal, reducing costs while improving the sensitivity; The gene fusion information at the RNA level has more direct clinical significance. Therefore, Nanodigmbio takes into account both the breadth and sensitivity of analysis, and has designed and developed a concentrated 105-gene panel specifically for RNA fusion, OncoFu Elite (for RNA) Panel v1.0, based on the transcript cDNA sequences.
カタログの1ページ目を開く
OncoFu Elite (for RNA) Panel v1.0 targets 105 genes which are the most common or relevant to clinically significant fusions in solid tumor studies. The probes cover all transcript coding regions and some UTR regions involved in the occurrence of fusions which are officially included in RefSeq 109. The panel supports the enrichment of fusion, mutation, and gene expression information at the RNA level. PDGFRA PDGFRBf PPARG* PRKACA PRKCA PRKCB Figure 1. Capture performance of OncoFu Elite (for RNA) Panel v1.0 (OncoFu E). A. Typical capture sequencing data; B. Comparison of the relative...
カタログの2ページ目を開く
Figure 2. Fusion detection of FFPE fusion RNA standards by OncoFu Elite (for RNA) Panel v1.0. A. Comparison of OncoFu E targeted enrichment and RNAseq; B. Targeted enrichment performance of OncoFu E after multiple dilution of the standards. Libraries of 100 ng Seraseq® FFPE Tumor Fusion RNA v4 Reference Material were prepared using NadPrep Total RNA-To-DNA Module with NadPrep DNA Universal Library Preparation Kit Series, separately carried out the direct sequencing (RNAseq), rRNA-depleted RNAseq (NEBNext rRNA Depletion Kit v2), and RNA capture sequencing (OncoFu E). 1 Gb data of each...
カタログの3ページ目を開く