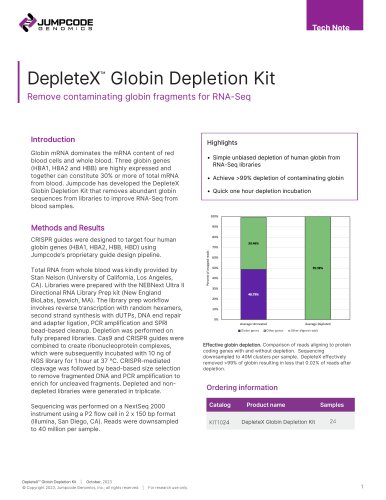
Catalog excerpts

Depleted" Single Cell RNA Boost KitRemove abundant and uninformative fragments prior to scRNA-Seq Highlights • Gain a deeper view of expression profiles of individual cells • Depletes sequences not used for secondary analysis including: Unaligned reads, ribosomal, mitochondrial, and optionally non-variable genes • Simple 3-step protocol integrated into any single cell sequencing workflow including 10x Genomics® Chromium™ Next GEM Single Cell 3' Introduction There are many opportunities to apply single-cell genomics to unanswered questions in biology. With its unique ability to study the individuality of cells, singlecell genomics has become an increasingly common, widely adopted approach. To profile large amounts of individual cells, transcriptional profiling with single-cell RNA Seq empowers researchers to know what genes are expressed, in what quantity, and how they differ across the cells within a sample. Because of this, single-cell studies require significant sequencing to understand transcript levels within individual cells. It can take anywhere from 50,000 to 150,000 reads per cell to sequence informative, lower abundance genes —with reads for a single sample often exceeding 150 million reads. Traditionally, single-cell data processing incorporates specific filtering and normalization steps prior to cell clustering and downstream interpretation. Instead of removing those reads in-silico, DepleteX Single Cell RNA Boost Kit removes those reads in-vitro before of sequencing, redistributing sequencing reads to unique biologically relevant transcripts. In this study, using PBMC cell types, we show an additional 2 cell types uncovered in the depleted condition compared to the control sample at the same sequencing depth. DepleteX leverages Cas9 and a specifically designed guide set to remove reads filtered by secondary analysis. DepleteX Single Cell RNA Boost Kit allows you to cut through the noise with minimal impact on your workflow and maximum confidence in your results. downstream for either short or long read technologies. Content for depletion was designed by analyzing a cohort of publicly available single cell 10x Genomics® data. By tailoring guides to deplete genomic intervals in addition to the highest expressed protein-coding ribosomal and mitochondrial genes, which are typically removed informatically downstream, we exhibited the ability to redistribute reads through in-silico depletion across samples representing 14 sample types. DepleteX™ Single Cell RNA Boost Kit | September, 2023 © Copyright 2023, Jumpcode Genomics, Inc.; all rights reserved | For research use only.
Open the catalog to page 1
Tech Note Methods PBMC samples were isolated from a donor and prepared using 10x Genomics® Chromium™ Next GEM Single Cell 3’ Reagent Kit (v3.1) protocol. In summary, barcoded PBMC cDNA from 3 individual channels were processed in parallel. For depleted samples, the barcoded cDNA from the same channels were processed simultaneously. Depletion was performed after adapter ligation. The 10X v3.1 protocol was resumed at step 3.5. All libraries were sequenced on a NovaseqTM 6000 to achieve a targeted read depth >30,000 reads per cell. We recovered ~9,000 cells per sample with each sample reaching...
Open the catalog to page 2
Finally, looking at Figure 3, the % of reads aligned to the genome, we have a 1.5X increase in the number of usable reads, or transcriptomic reads without ribosomal, mitochondrial and non-variable genes. DepleteX Single Cell RNA Boost Kit for 10x Genomics focuses on the signal, not the noise--to specifically remove uniformative sequences and improve biologically informative signal. Gain a deeper view of the expression profile of cells while still detecting the same number of genes with the DepleteX Single Cell RNA Boost Kit. Tcm/Naive helper T cells Classical monocytes Tem/Temra cytotoxic T...
Open the catalog to page 3All Jumpcode catalogs and technical brochures
-
DepleteX® Mito DNA Depletion Kit
11 Pages