 Website:
Leica Biosystems
Website:
Leica Biosystems
Group: Danaher
Catalog excerpts

Enhance Your Workflow with Automation Do you have a digital pathology slide scanner? xtract more data from your images with quantitative E image analysis Do you have large volumes of slides to score? utomatic batch image analysis works in the A background so you don‘t have to Do you worry about the accuracy of slide scoring? et quantitative, standardized data for a wide variety G of applications Do you find it time-consuming to maintain records of results? esults are automatically saved with digital R slide records and exported for further use Tumor and infiltrating lymphocytes in breast...
Open the catalog to page 2
Experience the Full Power of Digital Pathology with Aperio Image Analysis Unlimited assays provided by each algorithm ptimize & save flexible parameters to assist in O automating multiple assays Rapid set up & walk away protocol for batch analysis 5 clicks to analyze a batch of slides using your saved parameters Seamless workflow within Aperio Digital Pathology platform atch analysis of scanned slides supports whole slide B images & regions of interest straight from the scanner Wide range of applications & use cases nique & extensive outputs for each algorithm with U detailed color overlay...
Open the catalog to page 3
Aperio has a Flexible Menu of Image Analysis Algorithms Image Analysis RUO Menu peer-reviewed publications using Aperio Image Analysis Algorithms (PubMed database search for ‘Aperio’) Unlimited colors supported by algorithm Number of colors supported by algorithm in a single application A Key Tool for Many Applications in Published Biomedical Research For Research Use Only. Not for Use in Diagnostic Procedures.
Open the catalog to page 4
Aperio Image Analysis Algorithms at the Click of a Button COMPATIBLE STAINS Aperio GENIE • BRIGHTFIELD • FLUORESCENCE • Machine Learning • issue pattern recognition software T • rain to automatically identify tissue of interest in T digital images • ny chromogen, fluorochrome, counterstain, H&E, A special stain • xample: tumor regions of interest; kidney glomeruli; E pancreatic islets Aperio Membrane • BRIGHTFIELD • Antibody Assays • IHC of membrane antigens • rovides cell count for different intensity classes P • nly Brown membrane staining with Hematoxylin O counterstain • Example: HER2,...
Open the catalog to page 5
Aperio GENIE Algorithm Machine Learning Histology Pattern Recognition Aperio GENIE is an interactive image analysis tool for differentiating tissue subtypes within a digital slide. This Convoluted Neural Network (CNN) can be trained by the user with examples to automatically identify regions of interest for research, e.g. distinguishing tumor from normal tissue, or xenograft from native tissue. TISSUE CLASSIFICATION FOR BRIGHTFIELD AND FLUORESCENT IMAGES 1 Aperio Genie Classifier v1 1. Original digital image of H&E stained human breast tissue. 2. Aperio GENIE mask, with user-defined...
Open the catalog to page 6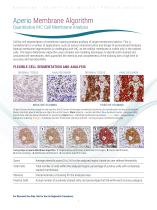
Aperio Membrane Algorithm Quantitative IHC Cell Membrane Analysis Cell-by-cell segmentation of membrane staining enables analysis of target membrane proteins. This is fundamental to a number of applications, such as cancer characterization and design of personalized therapies. Manual membrane segmentation is challenging with IHC, as the cellular membrane is visible only in the stained cells. The Aperio Membrane Algorithm uses complex cell modeling techniques to identify both stained and unstained cell membranes, then quantifies the intensity and completeness of the staining with a high...
Open the catalog to page 7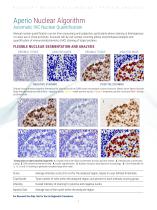
Aperio Nuclear Algorithm Automatic IHC Nuclear Quantification Manual nuclear quantification can be time-consuming and subjective, particularly where staining is heterogenous or nuclei are in close proximity. Accurate cell-by-cell nuclear counting allows morphological analysis and quantification of immunohistochemistry (IHC) staining of target proteins. FLEXIBLE NUCLEAR SEGMENTATION AND ANALYSIS ORIGINAL TISSUE ANALYSIS MASK ORIGINAL TISSUE NEGATIVE STAINING ANALYSIS MASK POSITIVE STAINING Original tissue showing negative (Hematoxylin) blue and positive (DAB) brown chromogen nuclear...
Open the catalog to page 8
Aperio Cytoplasmic Algorithm Automatic Positive and Negative Cytoplasm Quantification Manual analysis of complex IHC staining patterns involving nuclei and cytoplasm can be especially laborious when the nuclei are obscured by strong intensity staining. Automated and quantitative analysis of cellular staining is now possible for any IHC stain with the flexible Aperio Cytoplasmic Algorithm, producing results for both nuclear and cytoplasmic staining for a biomarker. QUANTITATIVE ANALYSIS OF CYTONUCLEAR STAINING ORIGINAL TISSUE ANALYSIS MASK Nucleus 3+ Nucleus 2+ Nucleus 1+ Nucleus 0+ STRONG...
Open the catalog to page 9
Automatic RNA In Situ Hybridization Quantification RNA ISH (Ribonucleic acid in situ hybridization) enables identification of individual copies of molecular targets within tissue, while maintaining morphology, a feature often lost in other methods such as PCR. Manual RNA ISH interpretation is time-consuming, subject to inter/intra-observer variability and typically employs semi-quantitative reads. The Aperio RNA ISH Algorithm enables accurate counting of individual signals across the tissue, providing standardized, reproducible results, including valuable per-cell data for export....
Open the catalog to page 10
Aperio Color Deconvolution Algorithm Separate and Analyze Chromogenic Stains The Aperio Color Deconvolution Algorithm separates a stained tissue image into multiple (up to 3) color channels, corresponding to the actual colors of the stains used. This enables the user to measure both the area and intensity of each stain across the tissue, even when the stains are superimposed at the same location. Final analysis masks are determined by the user: choose to display any 1 of the 3 separated colors or its corresponding intensity range. SEPARATE YOUR CHROMOGENS PIXEL BY PIXEL 1 1. Original...
Open the catalog to page 11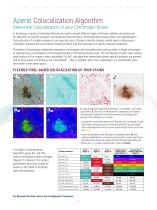
Aperio Colocalization Algorithm Determine Colocalization of your Chromogen Stains In histology, a variety of staining methods are used to target different types of tissues, cellular structures and for detection of specific proteins: conventional histochemistry, immunohistochemistry and in situ hybridization. Colocalization of multiple antigens is an important part of larger scientific studies, which seek to determine a correlation between the occurrence of these proteins and the outcome of a specific disease treatment. The Aperio Colocalization Algorithm separates chromogens and classifies...
Open the catalog to page 12All Leica Biosystems catalogs and technical brochures
-
APERIO VERSA
4 Pages
-
Aperio CS2
3 Pages
-
Aperio WebViewer DX User’s Guide
84 Pages
-
Aperio LIS Connectivity Overview
70 Pages
-
Aperio eSlide Manager
100 Pages
-
APERIO CLINICAL SOLUTION
7 Pages
-
Leica ASP6025 S
4 Pages
-
Leica VibratomeTM Series
8 Pages
-
Aperio GT 450 DX
5 Pages
-
BOND RXm
4 Pages
-
Leica St4020 Linear Stainer
2 Pages
-
HistoCore SPECTRA ST Stainer
4 Pages
-
Leica CM1520 Cryostat
2 Pages
-
Leica SM2010 R
4 Pages
-
Vibrating Blade Microtomes
8 Pages
-
HistoCore PEARL
4 Pages
-
CEREBRO
8 Pages
-
HistoCore PELORIS 3
3 Pages
-
Leica CM3050 S
4 Pages
-
2019 HistoCore Microtomes
8 Pages
-
Leica ST5010 Autostainer X
8 Pages
-
Leica CM1860/CM1860 UV
5 Pages
-
Aperio eSlide Manager
4 Pages
-
Leica CM1950
12 Pages
-
HistoCore BIOCUT
2 Pages
-
HistoCore Microtomes
4 Pages
-
Stereotaxic Solutions
7 Pages
-
HistoCore PERMA S
4 Pages
-
Leica CM3600 XP
2 Pages
-
Leica RM2125 RTS
2 Pages
-
Leica TP1020
4 Pages
-
Leica ASP300 S
8 Pages
-
Leica IP C
12 Pages
-
Cognitive Cxi
2 Pages













































