Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

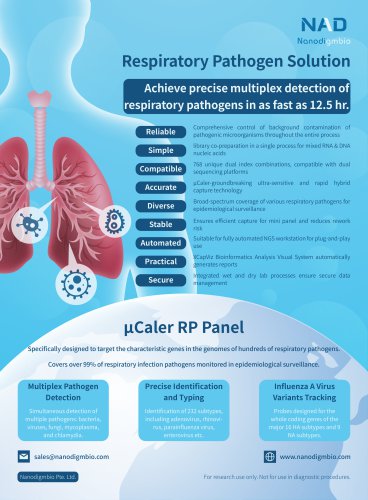
Acute Myeloid Leukemia (AML) accounts for 25.3% of all leukemias, and it is a clonal disorder characterized by the excessive proliferation of hematopoietic cells in bone marrow, leading to the rapid growth of abnormal cells in the bone marrow and blood, thereby interfering with hematopoiesis. Studies conducted both domestically and internationally have shown that Minimal Residual Disease (MRD) detection can be used for treatment response evaluation, relapse monitoring, treatment selection, and early intervention, making it a crucial step in reducing leukemia relapse and improving outcomes. The MRD detection methods for AML patients mainly include multiparameter flow cytometry (MFC), real-time quantitative PCR, digital PCR, and NGS. MFC is simple and fast but lacks the ability to determine specific leukemia subtypes at a low sensitivity. Real-time quantitative PCR and digital PCR have good specificity, high sensitivity, and are both relatively affordable, but they are only applicable to less than 40% of AML patients. On the other hand, NGS can simultaneously detect multiple mutation sites, observe clonal evolution, and facilitate high-throughput operations, making it to be considered as the "ultimate solution" applicable to all AML patients. However, NGS is associated with high sequencing costs and susceptibility to background noise, which needs to be overcome. Therefore, Nanodigmbio has designed and developed the ^Caler AML MRD comprehensive solution, which combines the exclusive patented |iCaler hybrid capture system with Unique Molecular Identifier (UMI). This solution enables ultra-high detection sensitivity and completes the entire experimental process within same day.
Open the catalog to page 1
m NadPrep UMI Adapter m UMI ■ NadPrep Universal UDI-Index Primer Mix Safe Stopping Point. ^Caler AML MRD comprehensive solution is based on liquid-phase hybridization target enrichment technology and specifically designed for adult Acute Myeloid Leukemia (AML) MRD research. This comprehensive solution selects MRD targets that cover approximately 90% of AML cases, allowing for the analysis of various mutations, including base substitutions, insertions/deletions, and gene fusions. By combining UMI and the |iCaler hybrid capture system, it achieves ultra-high detection sensitivity and can...
Open the catalog to page 2
• Precise coverage: ~90% of cases have mutations, with ~60% of cases having three or more mutations • Low background noise: Accurate and reliable with a high signal-to-noise ratio • Higher sensitivity: High conversion rate enables lower detection limits • Lower sequencing cost: >70% on-target rate, saving 80% of sequencing volume • Stable and efficient: Avoids inconsistent sequencing data and reduces rework • High-speed and convenient: Simplified experimental process, easy operation, and completion of the entire workflow within same day • Reference was made to multiple databases (COSMIC &...
Open the catalog to page 3
Lower sequencing cost Higher sensitivity Covaris Figure 2. Duplex depth (DCS211) detected by μCaler AML MRD compr- Figure 3. Minimum amount of raw data required for μCaler AML MRD fferent input conditions. The μCaler AML MRD comprehensive solution user to achieve a specific depth (satisfying Duplex Consensus Sequences ehensive solution and sonication-based library preparation under di- manual was used as a reference, and the filtering was performed based on Duplex Consensus Sequences (DCS211). The sequencing mode is Illumina Novaseq 6000, PE150. comprehensive solution and traditional...
Open the catalog to page 4
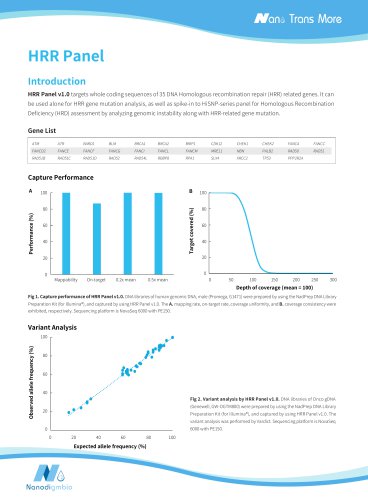
TraditionalpCalerDuration Traditional pCaler | Vacuum concentration Capture performanceA B ■Average depth (rmdup) SSCS DCS211 Mappability On-target 0.2x mean 0.5x mean Depth of coverage (mean=100) Figure 5. Capture performance of pCaler AML MRD comprehensive solution. 300 ng of fragmented DNA were used to prepare the pre-library, and the hybridization capture was completed with pCaler AML Panel v1.0. A. Mappability, On-target and target covered; B. Sequencing depth; C. Coverage uniformity and consistency. Note: The samples were derived from the proportionate mixing of Pancancer Light 800...
Open the catalog to page 5
Analysis of mutation in standard The samples were derived from the proportionate mixing of PancancerLight 800 gDNA Reference Standard (Genewell, GW-OG- TM 800) and Human Male Genomic DNA Standard (Promega, G1471) to simulate samples with different allele frequency (0.025% ~ 0.1%). The detection of different mutation sites in the simulated samples is as follows: A Reads supporting mutation Allele frequency Theoretical Figure 6. Mutation analysis of the μCaler AML MRD comprehensive solution. A. Average depth (DCS211 ≥ 1) B. Reads supporting mutations; C. Theoretical Repeat-1 Repeat-2 and...
Open the catalog to page 6
Ordering Information NEM Fragment Module Lib Prep Module Adapter Module Blocker Hybrid Capture Panel NadPrep DNA Library Preparation Kit (for Illumina®) G24 NadPrep NEM Fragment Module, 24 rxn NadPrep DNA Library Preparation Kit (for Illumina®) E96 NadPrep UMI Adapter Kit Set A1 (with 10 nt Index), 24 rxn NadPrep UMI Adapter Kit Set B1 (with 10 nt Index), 96 rxn NadPrep UMI Adapter Kit Set B2 (with 10 nt Index), 96 rxn μCaler NanoBlockers (for Illumina®), 16 rxn μCaler NanoBlockers (for Illumina®), 96 rxn μCaler Hybrid Capture Reagents, 16 rxn μCaler Hybrid Capture Reagents, 96 rxn μCaler...
Open the catalog to page 7All Nanodigmbio catalogs and technical brochures
-
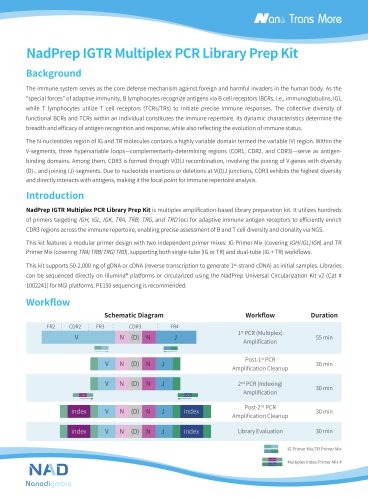
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages