Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

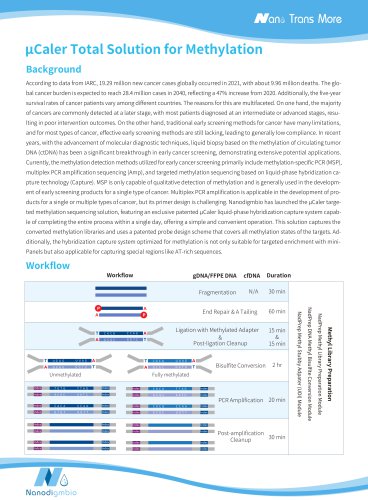
Ultra-sensitive Synchronous Detection of Methylation and Mutation | μCaler DNA Full Screen System Comprehensive Solution Background DNA methylation, as a chemical modification that does not alter the DNA sequence but can influence gene activity, plays a critical role in various biological processes, including gene expression regulation, embryonic development, cell proliferation and differentiation, and maintaining genomic stability. Abnormal DNA methylation is present throughout the entire process of cancer initiation and progression. Compared to traditional tumor markers, DNA methylation markers offer advantages such as earlier detection, less invasiveness, and greater precision. These markers are well-suited for early cancer diagnosis, risk assessment, minimal residual disease (MRD) monitoring, and guiding therapy. Currently, DNA methylation detection primarily relies on bisulfite (BS) conversion and enzymatic conversion techniques. BS con- version is fast and efficient, but the drastic changes in temperature and pH during the process can lead to DNA degradation and fragmentation, making it more suitable for samples with higher initial input amount. Enzymatic conversion is relatively mild but involves cumbersome and time-consuming procedures, with less stable conversion efficiency. DNA methylation detection based on NGS offers high throughput, making it suitable for the detection of single cancer type, multiple cancer types, or pan-cancer types. However, limited by the low initial amount of cfDNA samples and the need for simultaneous mutation detection, the operation of multi-omics analysis is complex and the high costs. To achieve the synchronous detection of methylation and mutation signals from a single, limited sample, Nanodigmbio has launched the ultra-sensitive solution for synchronous detection of methylation and mutation: μCaler DNA Full Screen System Comprehensive Solution. This solution only requires one initial sample, one hybridization reaction, and one set of capture probes to achieve an ultra-highly sensitive panoramic detection of methylation and mutation. μCaler DNA Full Screen System Comprehensive Solution integrates methylation sensitive restriction endonuclease (MSRE) technology with the exclusive patented μCaler Hybrid System, aiming to achieve ultra-high sensitivity in the synchronous det- ection of DNA methylation and mutation from a single sample. This solution utilizes MSRE technology, which specifically cleaves unmethylated sites; when a specific base within the recognition site is methylated, the cleavage is blocked. As a result, this app- roach can simultaneously generate both enzyme-digested and undigested methylated DNA libraries from the same sample. These libraries can then be used in the μCaler Hybrid System for the synchronous detection of DNA methylation and mutation within a single day. μCaler DNA Full Screen System Comprehensive Solution not only includes all the necessary reagents for the entire workflow but also offers customizable μCaler Full Screen Panel designs for target regions, as well as comprehensive bioinformatics analysis solutions. This fast, simple, user-friendly, and accurate end-to-end solution is designed to better support multi-omics resea
Open the catalog to page 1
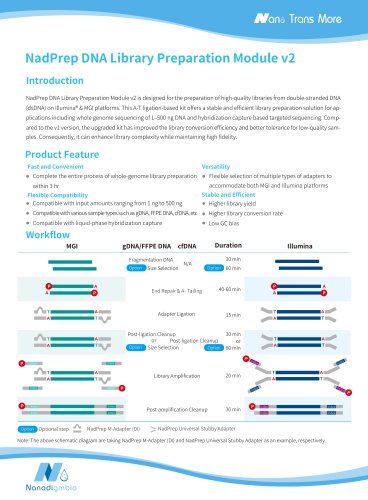
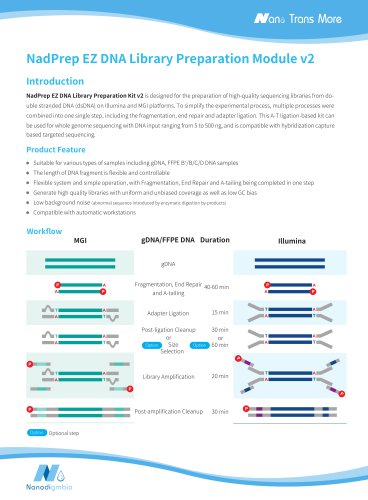
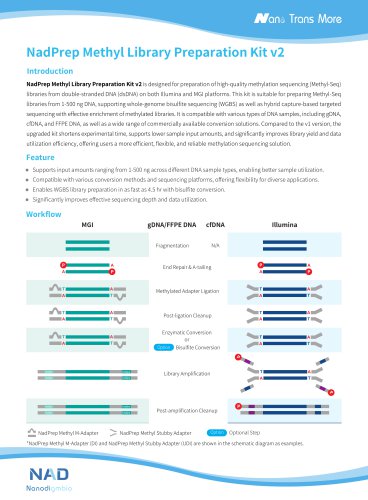
NadPrep DNA Library Preparation Kit v2 is designed for the preparation of high-quality libraries from double-stranded DNA (dsDNA) on Illumina® and MGI platforms. This A-T ligation-based kit offers a stable and efficient library preparation solution for applications including whole genome sequencing of 1‒500 ng DNA and hybridization capture based targeted sequencing. It has improved the library conversion efficiency and better tolerance for low-quality samples. Consequently, it can enhance library complexity while maintaining high fidelity. This kit is fully optimized for size selection with...
Open the catalog to page 2
cfDNA DNA Fragmentation and Quantification Quantification Index Index Index Index Index Index Index Index Index Index Post-amplification Cleanup Perform Hybridization Perform Capture and Elution NadPrep Universal Stubby Adapter Pre-library Amplification Perform Post-capture 30 min PCR & Purify and & Qutantify Library 30 min * The schematic diagram takes the NadPrep Universal Stubby Adapter as an example. μCaler Hybrid Capture MSRE Cleavage 80 min & & Post-cleavage Cleanup 30 min μCaler Hybrid Capture Reagents v2 μCaler NanoBlockers (for Illumina®) μCaler Full Screen Panel Adapter Ligation...
Open the catalog to page 3
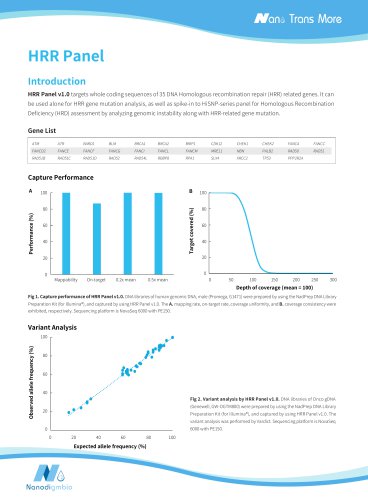
Stable and High Cleavage Efficiency A Cleavage efficiency of MSRE (%) Cleavage efficiency of MSRE (%) Simulated sample Figure 1. μCaler DNA Full Screen System can stably and efficiently cut the unmethylated sites across different sample types. A. Simulated samples with varying methylation levels and different initial input amounts of human genomic DNA standard (Promega, G1471); B. Clinical FFPE and cfDNA samples. Note: Samples were simulated DNA by using human 100% Methylated DNA standard (zymo, D5014-2) positive control and 0% Methylated DNA standard (zymo, D5014-1) negative control to...
Open the catalog to page 4
Accurate Quantification of Methylation Levels, Enhanced Detection Stability Full Screen Observed methylation level (%) Figure 3. Consistency of detection values obtained with the μCaler DNA Full Screen System: A. Detection values compared to theoretical values for samples with different methylation levels, and B. Consistency between two measurements of one randomly selected clinical colorectal cancer tissue sample. 1 M reads pair were randomly selected for data analysis. Note: Traditional_BS utilized NadPrep Hybrid Capture Reagents (overnight hybridization) for capture. Enhanced Sensitivity...
Open the catalog to page 5
Full Screen Reads supporting mutation Figure 4. Performance of μCaler DNA Full Screen System for synchronous detection of ultra-low-frequency mutation and low-abundance methylation. A total of 80 ng of simulated samples were analyzed using μCaler DNA Full Screen System with μCaler Full Screen LF Custom Panel (< 5 Kb, covering 31 CpG sites) for hybrid capture. 3 Gb were randomly selected for data analysis. A. Mutation frequencies at different sites corresponded closely with the nominal frequencies of the standard [pre-libraries containing UMI, with analytical filtering criteria set to duplex...
Open the catalog to page 6All Nanodigmbio catalogs and technical brochures
-
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages