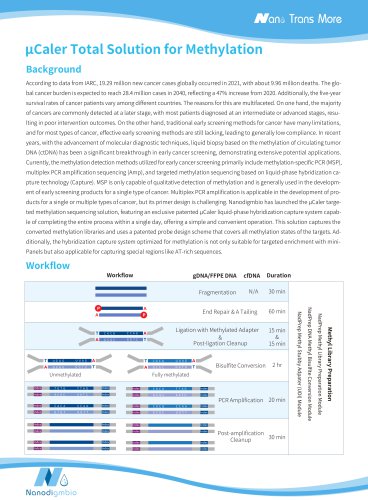
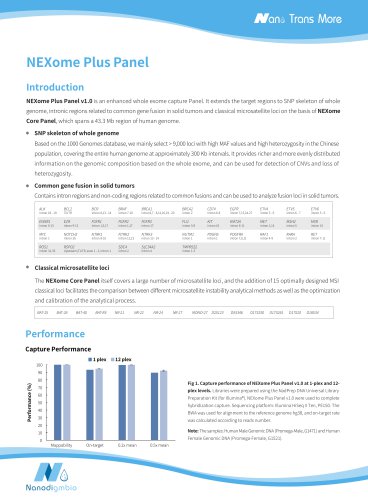
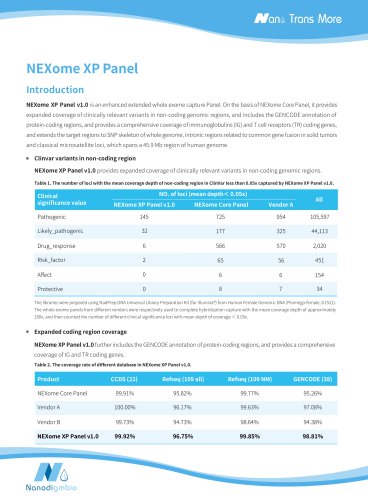
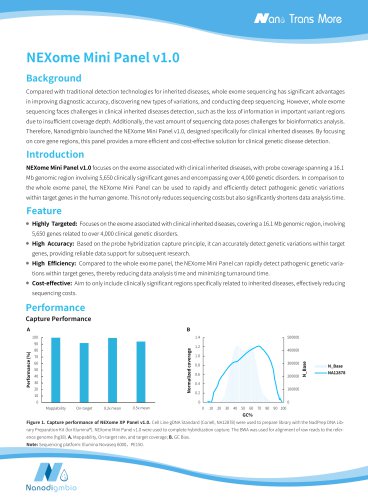
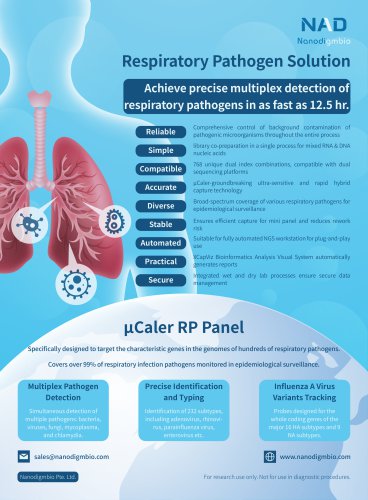
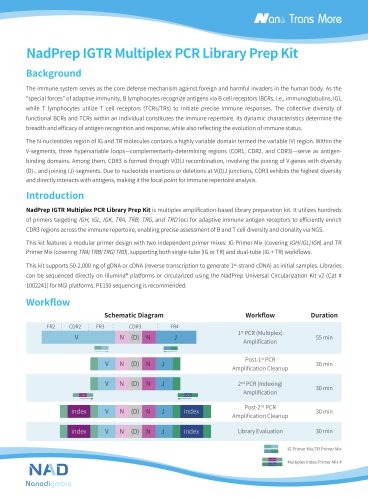
Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

Human leukocyte antigen (HLA), the major histocompatibility complex (MHC) of humans, is located on the short arm of chromosome 6 and is an important part of the human immune system. Different complexes bind to various exogenous or endogenous antigenic fragments through their binding grooves, which further interact with different T cells to activate downstream immune responses. Therefore, the functions undertaken by HLA place demands on its own diversity: HLA genes are among the most polymorphic region of human genome. HLA antigen polymorphisms are associated with a variety of diseases, as well as with vaccine and drug targeted population screening, tissue and organ transplantation. The compatibility of HLA matching is a critical factor for successful organ, bone marrow, and stem cell transplantation. In addition, HLA genotypes in cancer patients as well as systemic mutations in tumors may affect the efficacy of immunotherapy. Therefore, accurate typing of HLA genes is of great importance. HLAtyping Panel v1.0 targets a series of HLA genes and immune pathway genes, covering an approximately 40 Kb region of the genome. The Panel has been optimized for polymorphisms in the exonic region of the classical HLA gene to ensure balanced capture of alleles and support typing to the third field level. HLA-A HLA-B HLA-C HLAtyping Panel v2.0 refers to the latest database (Version 3.50), and targets 11 HLA genes (Class I: HLA-A, B, C; Class II: HLA-DPA1, DPB1, DQA1, DAQB1, DRB1/3/4/5). The probe covers the full-length genome sequence of all alleles and supports high-precision typing to the fourth field level.
Open the catalog to page 1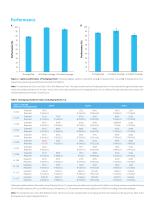
On-target Rate >0.2x Mean Coverage >0.5x Mean Coverage On-target Rate >0.2x Mean Coverage >0.5x Mean Coverage Figure 1. Capture performance of HLAtyping Panel. Following targeted capture of standards using A. HLAtyping Panel v1.0 and B. HLAtyping Panel v2.0, respectively, sequencing was performed using Illumina® platform. Note: The standards are Class I and Class II UCLA DNA Reference Panel. The capture performance of HLAtyping Panel v1.0 was reflected through calculated mean values and standard deviations of 24 Class I and 12 Class II; the capture performance of HLAtyping Panel v2.0 was...
Open the catalog to page 2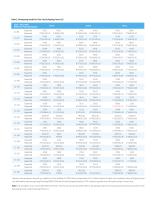
ClasslUCLA DNA Reference Panel (Standard) HLA-A Libraries were prepared using 50 ng of gDNA and the NadPrep EZ DNA Library Preparation Kit v2, hybrid capture (6 plex) was completed using HLAtyping Panel v2.0, followed by sequencing using NovaSeq 6000 PE150 with the data of approximately 0.5 Gb. Sequencing data were HLA typed using in-house tools. Note: The standard is Class I UCLA DNA Reference Panel. The first row of each standard refers to the typing result for the standard and the second row refers to the data typing result using HLAtyping Panel v2.0.
Open the catalog to page 3
Figure 2. Coverage of reference sequence under non-mismatching conditions with targeted capture for C1-213 using HLAtyping Panel v2.0. The sequence data for typing results was aligned to the reference sequence of the type. Individual results with differences in the second field level presented in the typing results of Class I using HLAtyping Panel v2.0. After sequence alignment and inspection, it was found that the new typing results using HLAtyping Panel v2.0 were more in line with the situation: The whole reference sequence showed continuous coverage under non-mismatching conditions;...
Open the catalog to page 4All Nanodigmbio catalogs and technical brochures
-
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages