Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

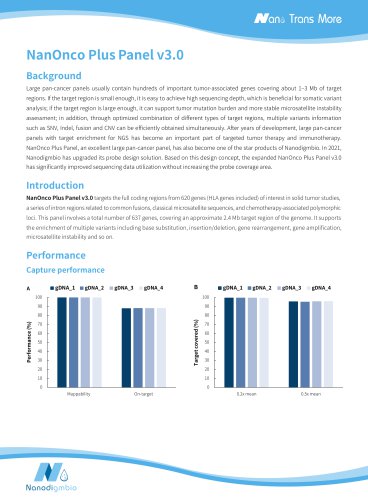
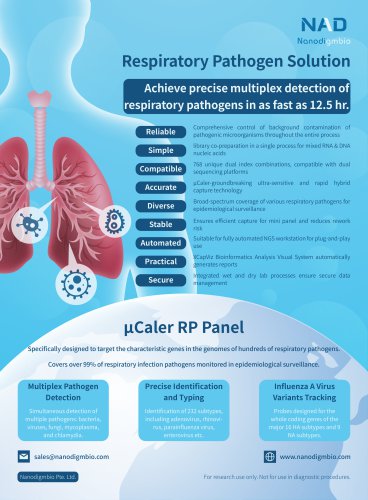
NanOnco Plus Panel v3.0 Background Large pan-cancer panels usually contain hundreds of important tumor-associated genes covering about 1‒3 Mb of target regions. If the target region is small enough, it is easy to achieve high sequencing depth, which is beneficial for somatic variant analysis; if the target region is large enough, it can support tumor mutation burden and more stable microsatellite instability assessment; in addition, through optimized combination of different types of target regions, multiple variants information such as SNV, Indel, fusion and CNV can be efficiently obtained simultaneously. After years of development, large pan-cancer panels with target enrichment for NGS has become an important part of targeted tumor therapy and immunotherapy. NanOnco Plus Panel, an excellent large pan-cancer panel, has also become one of the star products of Nanodigmbio. In 2021, Nanodigmbio has upgraded its probe design solution. Based on this design concept, the expanded NanOnco Plus Panel v3.0 has significantly improved sequencing data utilization without increasing the probe coverage area. NanOnco Plus Panel v3.0 targets the full coding regions from 620 genes (HLA genes included) of interest in solid tumor studies, a series of intron regions related to common fusions, classical microsatellite sequences, and chemotherapy-associated polymorphic loci. This panel involves a total number of 637 genes, covering an approximate 2.4 Mb target region of the genome. It supports the enrichment of multiple variants including base substitution, insertion/deletion, gene rearrangement, gene amplification, microsatellite instability and so on. Target covered (%)
Open the catalog to page 1
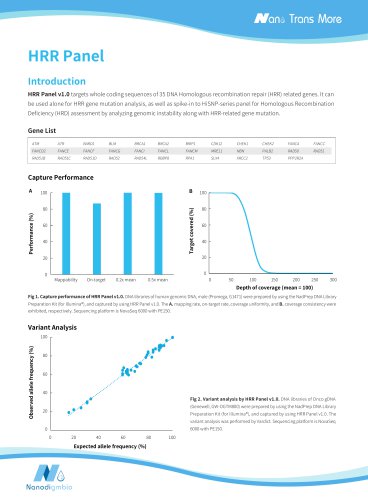
Target covraed (%) libraries. Libraries were prepared using NadPrep EZ DNA Library Preparation Kit coupled with NadPrep UDI Adapter (for Illumina®), and sequenced on Illumina Novaseq 6000 with PE150. The BWA was used for alignment to the reference genome hg38 and on-target rate was calculated by the number of reads. A. Mappability and on-target rate; B. Target covered; C. Coverage uniformity and Note: The gDNA_1-4 samples were: Human Male Genomic DNA (Promega- male, G1471); Onco Structural Multplex 5% gDNA (Genewell, GW-OGTM001); Fig 1. Capture performance of NanOnco Plus Panel v3.0 in...
Open the catalog to page 2
Table 1. Detection of other variant types using NanOnco Plus Panel v3.0. Expected allele frequency (%) / copy number Reference observed (%) Wild type observed (%) CNV MET Amplification 3.5 copies 3.4 copies 2 copies CNV ERBB2 Amplification 7.0 copies 8.3 copies 2 copies Libraries were prepared using NadPrep EZ DNA Library Prepation Kit coupled with NadPrep UDI Adapter (for Illumina®), and sequenced on Illumina Novaseq 6000 with PE150. Variant analysis were performed using Vardict, Delly and CNVkit. The average sequencing depth of GW-OGTM001 and GW-OGTM005 is 966.71x and 1,126.04x,...
Open the catalog to page 3
RPTOR RRAGC RRAS RRAS2 RSPO2 RTEL1 PTPRO PTPRS PTPRT QKI RAB35 Selected intronic regions intron 9-12 intron 1,5,17 intron 1,17 intron 26 intron 8-10 intron 12,15 Microstatellite markers Medicine related sites SNPs Ordering Information Product NanOnco Plus Panel v3.0, 16 rxn NanOnco Plus Panel v3.0, 96 rxn 100m2F For research use only. Not for use in diagnostic procedures. Versionl.1 Nanodigmbio Pte. Ltd.Service: sales@nanodigmbio.com | Web: www.nanodigmbio.com Nanodigmbio Pte. Ltd. reserves all rights. H
Open the catalog to page 4All Nanodigmbio catalogs and technical brochures
-
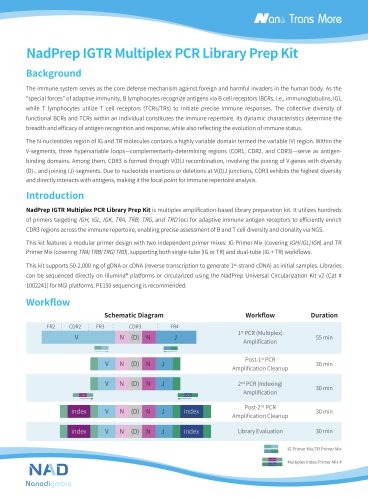
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages