Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

NEXome Core Panel Whole-exome sequencing is a widely used next-generation sequencing (NGS) method. The human exome represents less than 2% of the genome, but contains ~85% of known disease-related variants. Not only that, the exome is distributed throughout the genome, and it also covers tens of thousands of high-frequency SNP sites and nearly 20,000 microsatellite sites. Therefore, whole exome sequencing (WES) is not only used in genetic disease screening, tumor mutation burden and other sequence variation analysis,but also used in copy number variation, homologousre combination repair deficiency detection and other structural variation analysis and microsatellite instability analysis, etc. Therefore, based on the latest version of Refseq (109, 2021) and the CCDS database combined with individual requirements, Nanodigmbio has independently developed a core version of the all-exome gene detection panel: NEXome Core Panel. On the basis of the latest version of Refseq (109, 2021) and the CCDS database, NEXome Core Panel has carefully selected some regions that are worthy of attention outside the database. At the same time, based on product continuity considerations, some regions deleted in the latest version of the database have been retained. The NEXome Core Panel contains approximately 400,000 single-stranded DNA probes that have been independently synthesized and independently inspected for quality, tar- geting a 34.7Mb genomic region (19,613 genes). NEXome Core Panel, as a core all-exome Panel, can be combined with differ- ent spike-in panels to meet different application requirements. Production and Quality Control Independently Synthesized and Independently Inspected for Quality 37538.6 Table 1. Examples for analysis detection of each probe. Fig 1. Mass spectrometry results of a single probe from NEXome Core Panel. The theoretical molecular weight was 37542.9 Da, and the mass spectrometry results showed that the measured molecular weight was 37538.6 Da. Extinction Calculated Measured M.W. coeff
Open the catalog to page 1
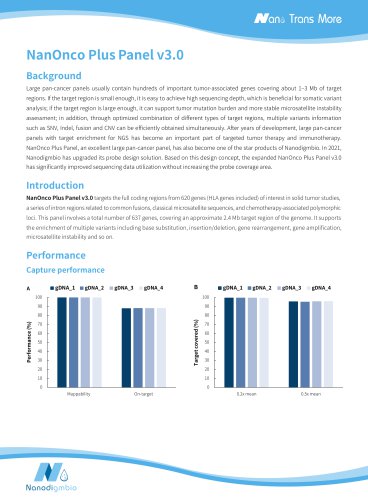
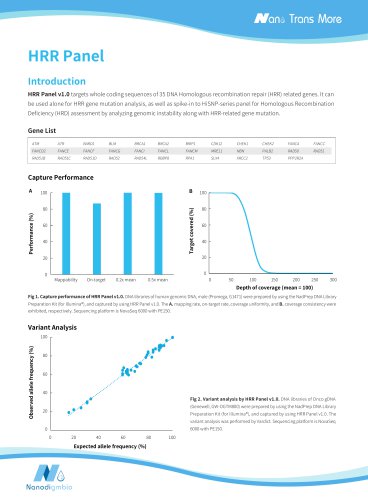
Target covered (%) Depth of coverage (mean = 100) Normalized coverage Target covered (%) Fig 2. Capture performance of the NEXome Core Panel in different gDNA libraries. A. Mappability and On-target rate, B. Target covered, C. Coverage uniformity and consistency, D. The unbiased of GC coverage; DNA libraries were prepared using the NadPrep DNA Universal Library Preparation Kit Series, NEXome Core Panel (6 plex) were respectively used to complete hybridization capture. The BWA was used for alignment of raw reads to the reference genome (hg38). Sequencing platform: Illumina Novaseq...
Open the catalog to page 2
Product Advantage On-target Fold 80 base penalty Depth/M read pair Fig 4. Performance comparison of different whole exome panel for Fig 5.The achievable coverage depth of target area for different whole for alignment of raw reads to the reference genome (hg38). Sequencing used for alignment of raw reads to the reference genome (hg38). Select all data on-target rate and target covered of gDNA libraries. The BWA was used platform: Illumina Novaseq 6000,PE150. Ordering Information exome panel under the same amount of sequencing data. The BWA was to analyze according to every M read pair....
Open the catalog to page 3All Nanodigmbio catalogs and technical brochures
-
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages