Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

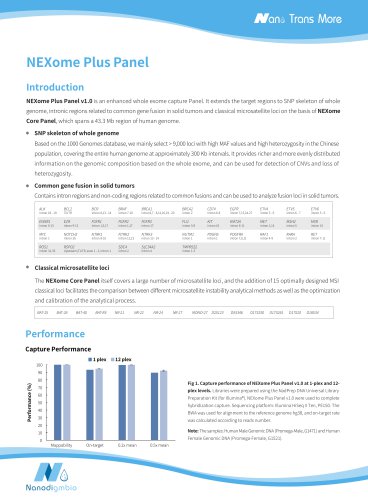
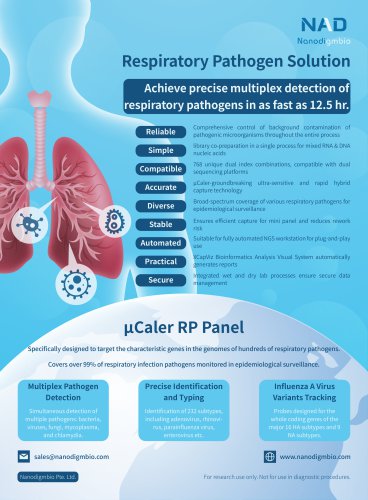
NEXome Plus PanelIntroduction NEXome Plus Panel v1.0 is an enhanced whole exome capture Panel. It extends the target regions to SNP skeleton of whole genome, intronic regions related to common gene fusion in solid tumors and classical microsatellite loci on the basis of NEXome Core Panel, which spans a 43.3 Mb region of human genome. Based on the 1000 Genomes database, we mainly select > 9,000 loci with high MAF values and high heterozygosity in the Chinese population, covering the entire human genome at approximately 300 Kb intervals. It provides richer and more evenly distributed information on the genomic composition based on the whole exome, and can be used for detection of CNVs and loss of heterozygosity. Contains intron regions and non-coding regions related to common fusions and can be used to analyze fusion loci in solid tumors. intron 9-12 intron 1,5,17 intron 1,17 intron 26 intron 8-10 intron 12,15 intron 16 intron 6-11 intron 1,14 intron 1 intron 7,9,11 intron 4-9 • Classical microsatellite lociThe NEXome Core Panel itself covers a large number of microsatellite loci, and the addition of 15 optimally designed MSI classical loci facilitates the comparison between different microsatellite instability analytical methods as well as the optimization and calibration of the analytical process. BAT-25 BAT-26 BAT-40 BAT-RII NR-21 NR-22 NR-24 NR-27 MONO-27 D2S123 D5S346 D17S250 D17S261 D17S20 D18S34 Capture Performance Fig 1. Capture performance of NEXome Plus Panel v1.0 at 1-plex and 12-plex levels. Libraries were prepared using the NadPrep DNA Universal Library Preparation Kit (for Illumina®). NEXome Pius Panel v1.0 were used to complete hybridization capture. Sequencing platform: Illumina HiSeq X Ten, PE150. The BWA was used for alignment to the reference genome hg38, and on-target rate was calculated according to reads number.
Open the catalog to page 1
Target covered (%) Depth of coverage (mean = 100) Target covered (%) Fig 2. Capture performance of NEXome Plus Panel v1.0 in different gDNA libraries. Libraries were prepared using the NadPrep DNA Universal Library Construction Kit (for Illumina®). NEXome Plus Panel v1.0 (12 plex) were used to complete hybridization capture. Ssequencing platform: Illumina HiSeq X Ten with PE150. The BWA was used for alignment to the reference genome hg38, and on-target rate was calculated according to reads number. A. Mappability and on-target rate; B. Target covered; C. Coverage uniformity and consistency;...
Open the catalog to page 2All Nanodigmbio catalogs and technical brochures
-
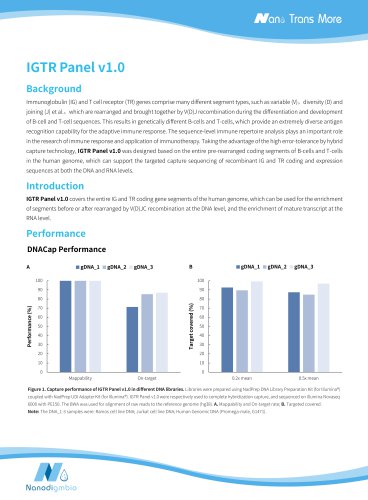
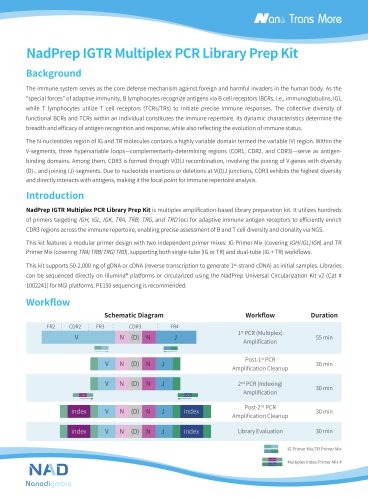
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages