Website:
Nanodigmbio
Website:
Nanodigmbio
Catalog excerpts

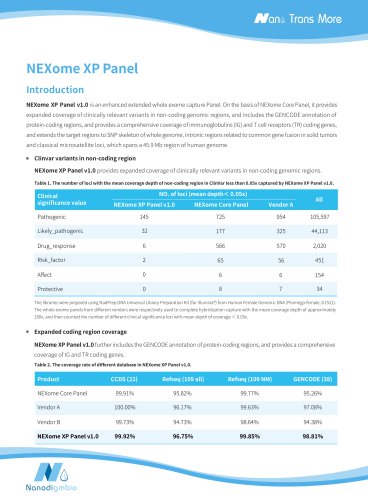
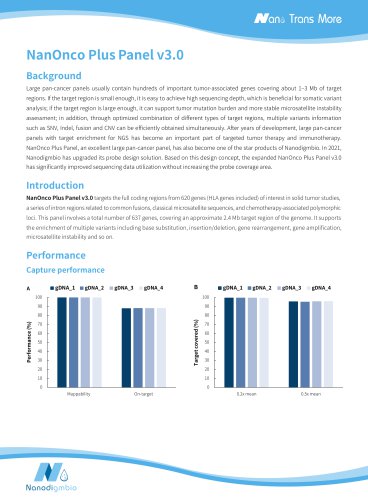
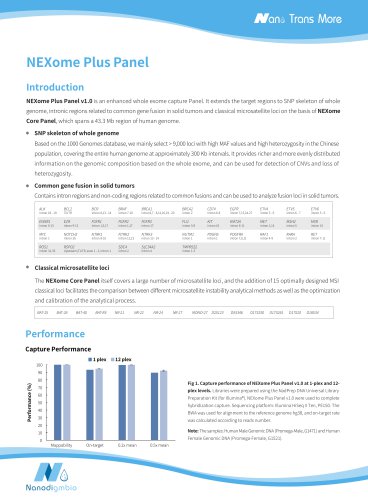
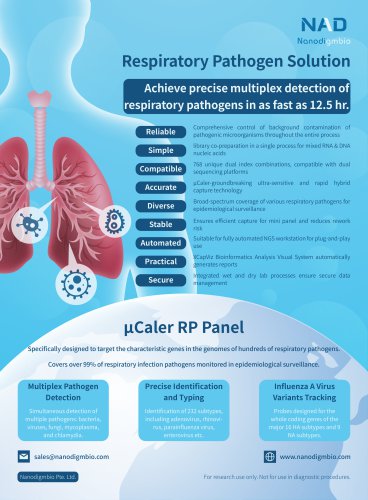
NEXome XP PanelIntroduction NEXome XP Panel v1.0 is an enhanced extended whole exome capture Panel. On the basis of NEXome Core Panel, it provides expanded coverage of clinically relevant variants in non-coding genomic regions, and includes the GENCODE annotation of protein-coding regions, and provides a comprehensive coverage of immunoglobulins (IG) and T cell receptors (TR) coding genes, and extends the target regions to SNP skeleton of whole genome, intronic regions related to common gene fusion in solid tumors and classical microsatellite loci, which spans a 45.9 Mb region of human genome. NEXome XP Panel v1.0 provides expanded coverage of clinically relevant variants in non-coding genomic regions. Table 1. The number of loci with the mean coverage depth of non-coding region in ClinVar less than 0.05x captured by NEXome XP Panel v1.0. Clinical NO. of loci (mean depthC 0.05x) significance value NEXome XP Panel v1.0 NEXome Core Panel Vendor A The libraries were prepared using NadPrep DNA Universal Library Preparation Kit (for Illumina®) from Human Female Genomic DNA (Promega-female, G1521). The whole exome panels from different vendors were respectively used to complete hybridization capture with the mean coverage depth of approximately 100x, and then counted the number of different clinical significance loci with mean depth of coverage < 0.05x. NEXome XP Panel v1.0 further includes the GENCODE annotation of protein-coding regions, and provides a comprehensive coverage of IG and TR coding genes. Table 2. The coverage rate of different database in NEXome XP Panel v1.0. Product CCDS (22) Refseq (109 all) Refseq (109 NM) GENCODE (38) NEXome Core Panel 99.91% 95.82%
Open the catalog to page 1
• SNP skeleton of whole genome Based on the 1000 Genomes database, we mainly select > 9,000 loci with high MAF values and high heterozygosity in the Chinese population, covering the entire human genome at approximately 300 Kb intervals. It provides richer and more evenly distributed information on the genomic composition based on the whole exome, and can be used for detection of CNVs and loss of heterozygosity. • Common gene fusion in solid tumors Contains intron regions and non-coding regions related to common fusions and can be used to analyze fusion loci in solid tumors. intron 6-13...
Open the catalog to page 2
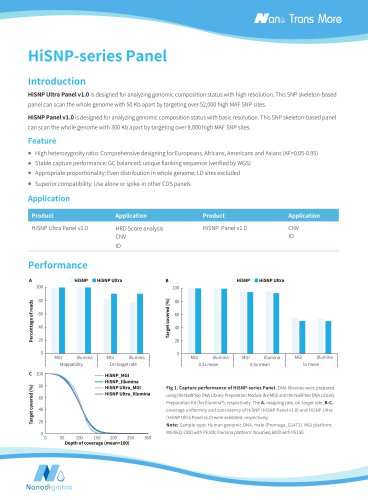
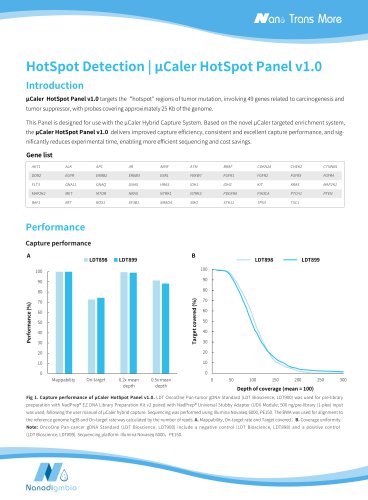
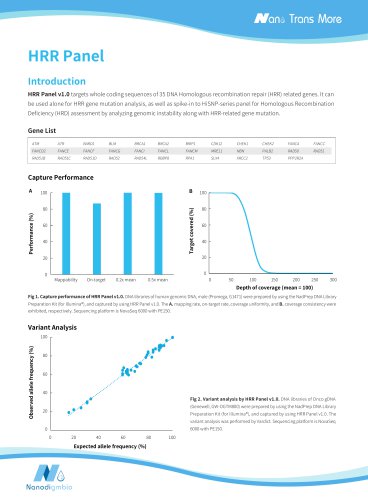
Target covered (%) Depth of coverage (mean = 100) Normalized coverage Fig 1. Capture performance of NEXome XP Panel v1.0 in different gDNA libraries. The libraries were prepared using NadPrep DNA Universal Library Preparation Module Series coupled with NadPrep Universal Stubby Adapter (UDI) Module and NadPrep Universal Adapter (MDI) Module (for MGI) from different gDNA. NEXome XP Panel v1.0 were respectively used to complete hybridization capture. The BWA was used for alignment of raw reads to the reference genome (hg38). A. Mappability and On-target rate; B. Targeted covered; C. Coverage...
Open the catalog to page 3
High Confidence Variant Calls A Indels calling rate (%) SNPs calling rate (%) Target region Target region Fig 3. Sensitivity and positive predictive value (PPV) of NEXome XP Panel v1.0 for NA12878 mutation identification. A. SNPs;B. Indels. Note: Sample type: Cell Line gDNA Standard (Coriell, NA12878). Sequencing platform: Illumina Novaseq 6000,PE150;DNBSEQ-G400,PE150. Ordering Information Product NEXome XP Panel v1.0, 16 rxn NEXome XP Panel v1.0, 96 rxn For research use only. Not for use in diagnostic procedures. Without the written permission of Nanodigmbio, no other individual or...
Open the catalog to page 4All Nanodigmbio catalogs and technical brochures
-
Nanodigmbio- IGTR Panel v1.0
4 Pages
-
Nanodigmbio-HGBP Panel v1.0
3 Pages
-
Nanodigmbio-NanOnCT Panel
2 Pages
-
Nanodigmbio-NGS HRR Panel
2 Pages